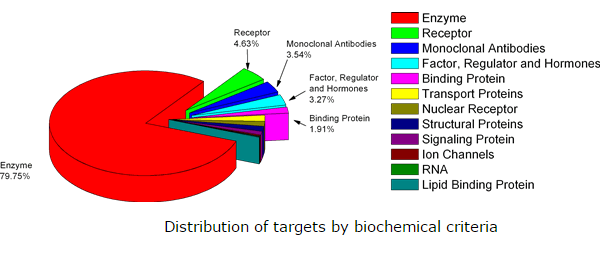
PDTD is a web-accessible protein database for in silico target identification. It currently contains more than 1100 protein entries with 3D structures presented in the Protein Data Bank. The data are extracted from the literatures and several online database such as TTD, DrugBank and Thomson Pharma. The database covers diverse information of more than 830 known or potential drug targets, including protein and active sites structure in both PDB and mol2 formats, related diseases, biological functions as well as associated regulating pathways. Each target is categorized by both nosology and biochemical function. PDTD supports keyword search function, such as PDB ID, target name, and disease name. Data set generated by PDTD can be viewed with the plug-in of molecular visualization tools and also can be downloaded freely. Remarkably, PDTD is specially designed for target identification. In conjunction with TarFisDock, PDTD can be used to identify binding proteins for small molecules. The results can be downloaded in the form of mol2 file with the binding pose of the probe compound and a list of potential binding targets according to their ranking scores.
PDTD serves as a comprehensive and unique repository of drug targets. Integrated with TarFisDock, PDTD is a useful resource to identify binding proteins for active compounds or existing drugs. Its potential applications include in silico drug target identification, virtual screening, and the discovery of the secondary effects of an old drug (i.e. new pharmacological usage) or an existing target (i.e. new pharmacological or toxic relevance), thus it may be a valuable platform for the pharmaceutical researchers. PDTD is available online at http://www.dddc.ac.cn/pdtd/

No comments:
Post a Comment